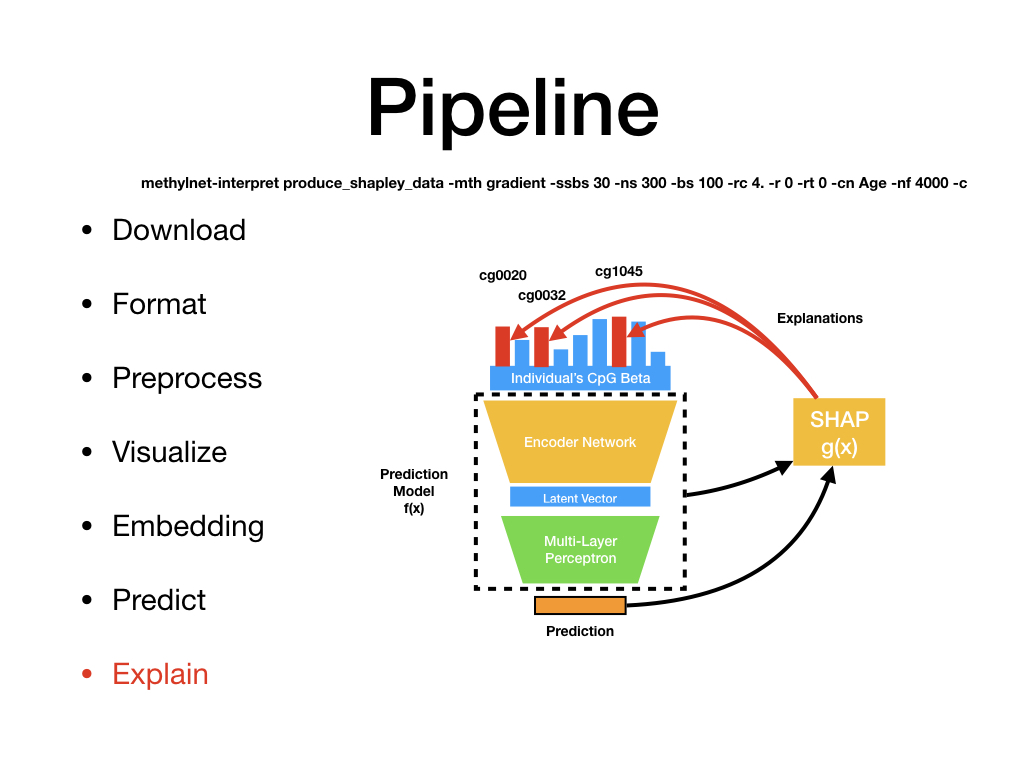
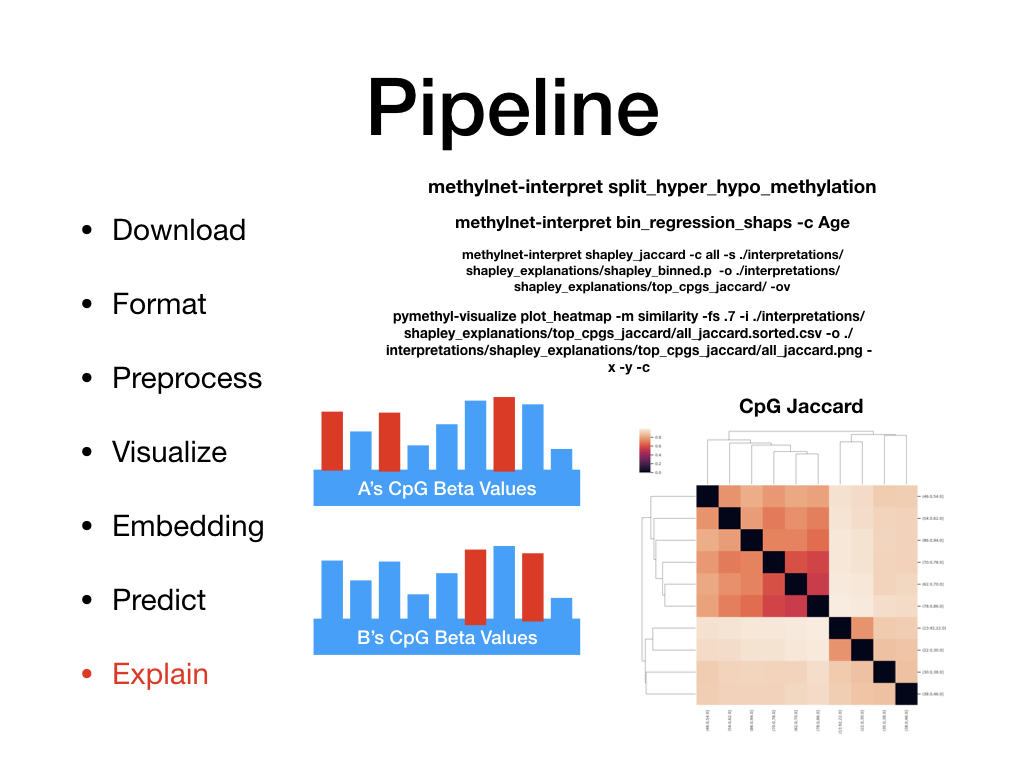
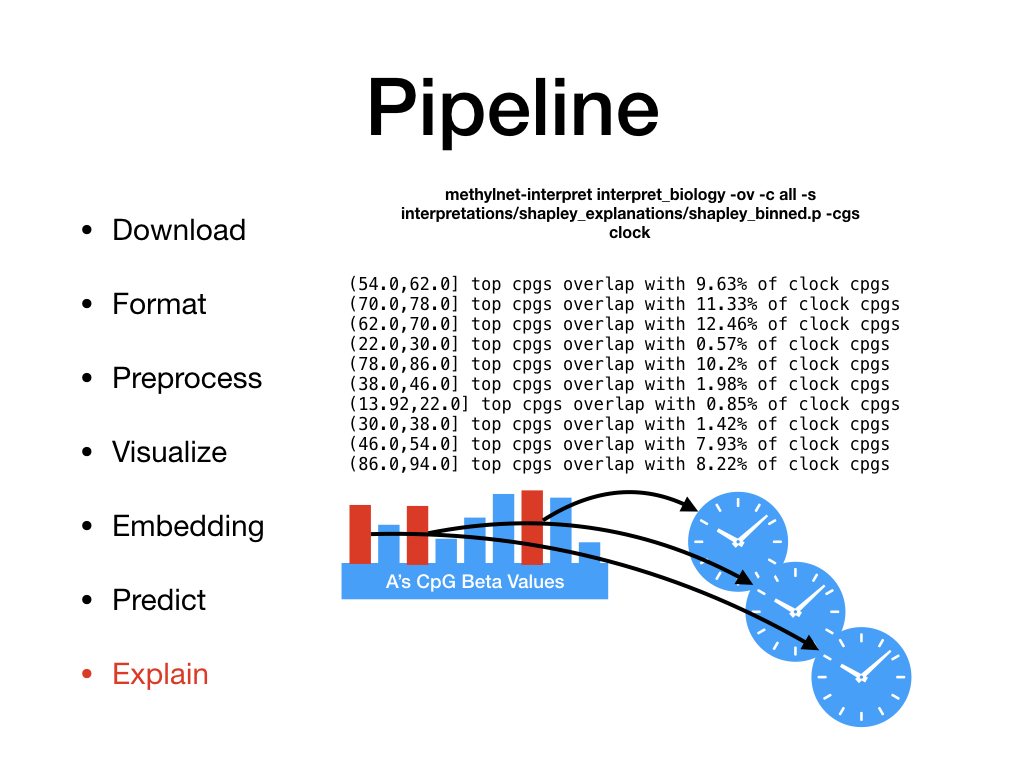
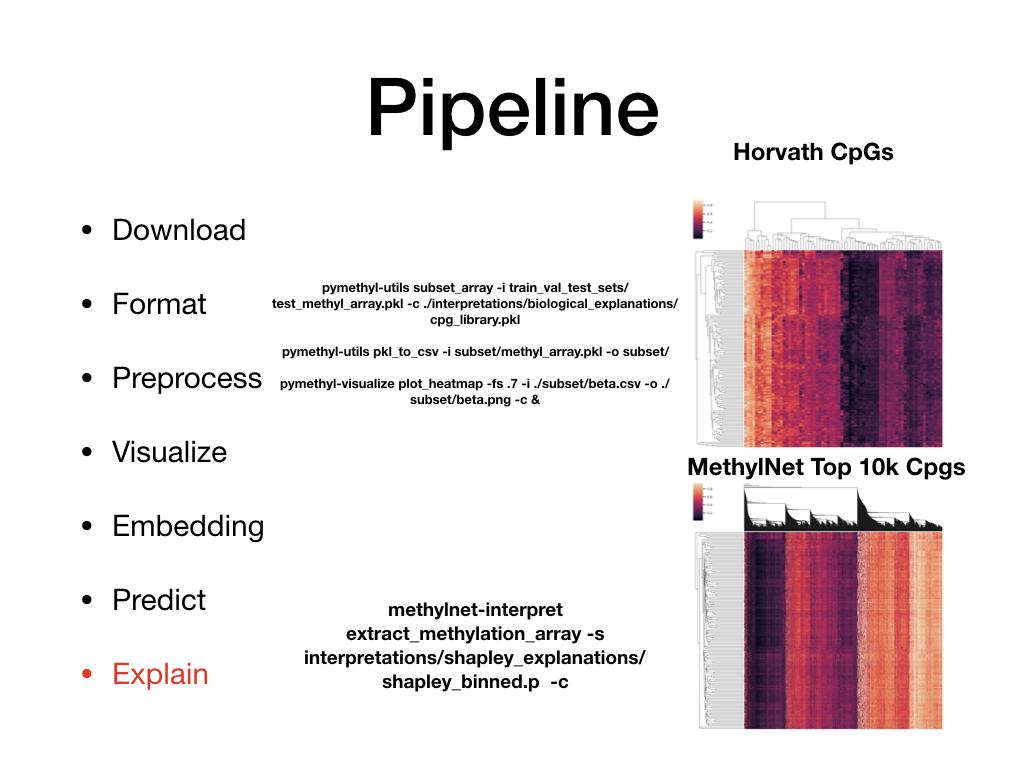
Welcome to MethylNet’s documentation!¶
https://github.com/Christensen-Lab-Dartmouth/MethylNet
See README.md in Github repository for install directions and for example scripts for running the pipeline (not all datasets may be available on GEO at this time).
There is both an API and CLI available for use. Examples for CLI usage can be found in ./example_scripts.
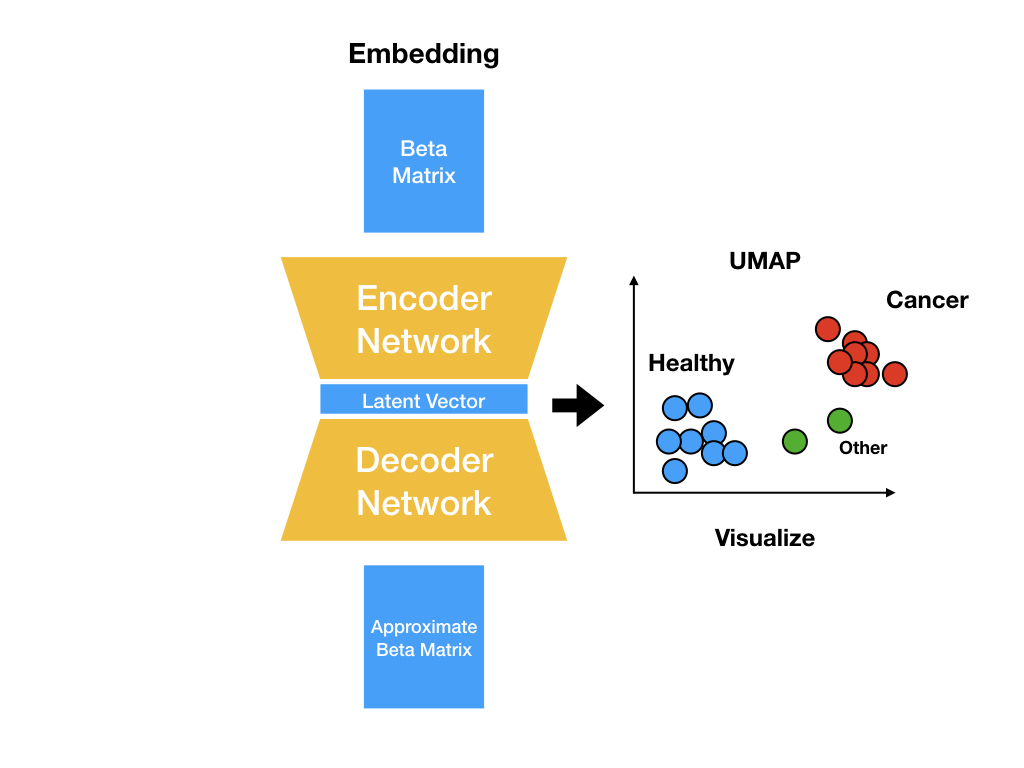
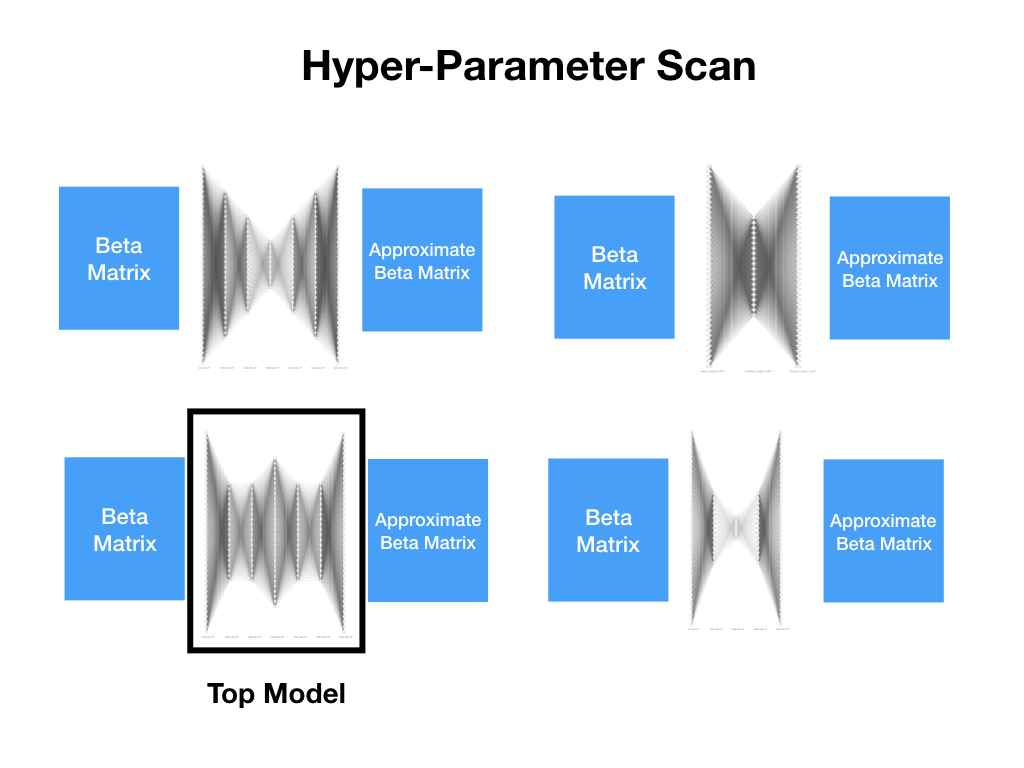
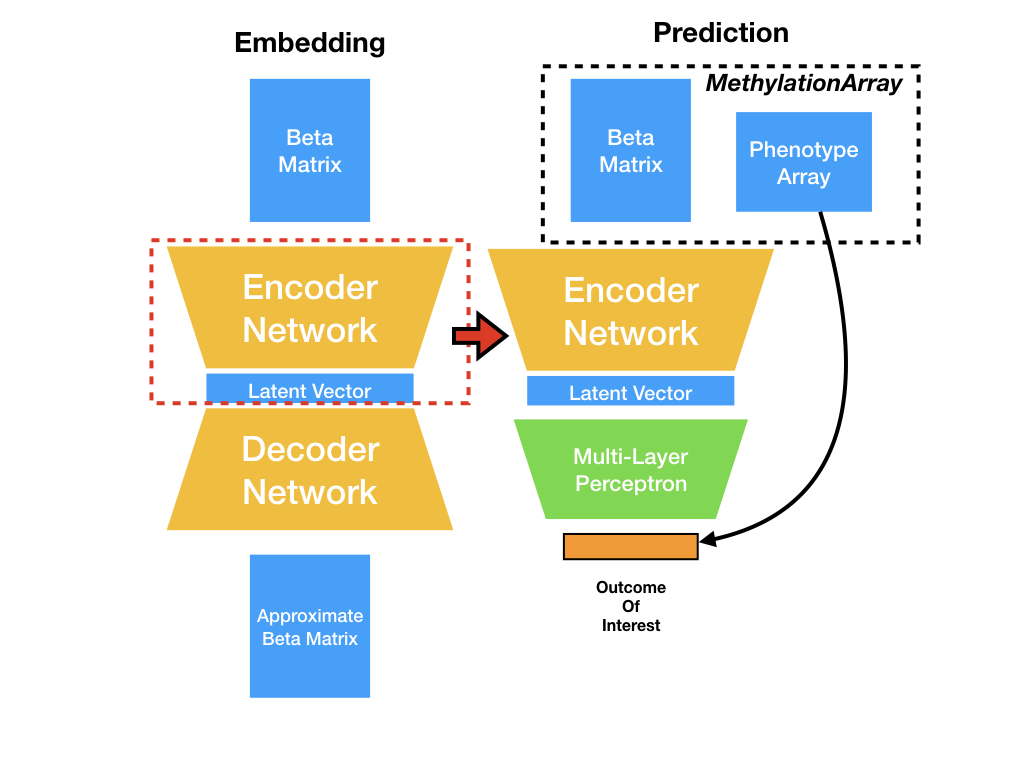
datasets.py¶
Contains datatypes core to loading MethylationArrays into Torch tensors.
-
class
methylnet.datasets.MethylationDataSet(methylation_array, transform, outcome_col='', categorical=False, categorical_encoder=False)[source]¶ Pytorch Dataset that contains instances of methylation array samples to be loaded.
Parameters: - methylation_array : MethylationArray
Methylation Array input.
- transform : Transformer
Transforms data into torch tensor.
- outcome_col : str
Pheno column(s) to train on.
- categorical : bool
Whether predicting categorical/classification.
- categorical_encoder : encoder
Encoder used to binarize categorical column.
Attributes: - samples : np.array
Samples of MethylationArray
- features : np.array
List CpGs.
- encoder :
Encoder used to binarize categorical column.
- new_shape :
Shape of torch tensors.
- length :
Number CpGs.
- methylation_array
- outcome_col
- transform
Methods
to_methyl_array(self)Convert torch dataset back into methylation array, useful because turning into torch dataset can cause the original MethylationArray beta matrix to turn into numpy array, when needs turn back into pandas dataframe.
-
class
methylnet.datasets.MethylationGenerationDataSet(methylation_array, transform, outcome_col='', categorical=False, categorical_encoder=False)[source]¶ MethylationArray Torch Dataset that contains instances of methylation array samples to be loaded, specifically targetted for prediction.
Parameters: - methylation_array : MethylationArray
Methylation Array input.
- transform : Transformer
Transforms data into torch tensor.
- outcome_col : str
Pheno column(s) to train on.
- categorical : bool
Whether predicting categorical/classification.
- categorical_encoder : encoder
Encoder used to binarize categorical column.
Attributes: - samples : np.array
Samples of MethylationArray
- features : np.array
List CpGs.
- encoder :
Encoder used to binarize categorical column.
- new_shape :
Shape of torch tensors.
- length :
Number CpGs.
- methylation_array
- outcome_col
- transform
Methods
to_methyl_array(self)Convert torch dataset back into methylation array, useful because turning into torch dataset can cause the original MethylationArray beta matrix to turn into numpy array, when needs turn back into pandas dataframe.
-
class
methylnet.datasets.MethylationPredictionDataSet(methylation_array, transform, outcome_col='', categorical=False, categorical_encoder=False)[source]¶ MethylationArray Torch Dataset that contains instances of methylation array samples to be loaded, specifically targetted for prediction.
Parameters: - methylation_array : MethylationArray
Methylation Array input.
- transform : Transformer
Transforms data into torch tensor.
- outcome_col : str
Pheno column(s) to train on.
- categorical : bool
Whether predicting categorical/classification.
- categorical_encoder : encoder
Encoder used to binarize categorical column.
Attributes: - samples : np.array
Samples of MethylationArray
- features : np.array
List CpGs.
- encoder :
Encoder used to binarize categorical column.
- new_shape :
Shape of torch tensors.
- length :
Number CpGs.
- methylation_array
- outcome_col
- transform
Methods
to_methyl_array(self)Convert torch dataset back into methylation array, useful because turning into torch dataset can cause the original MethylationArray beta matrix to turn into numpy array, when needs turn back into pandas dataframe.
-
class
methylnet.datasets.RawBetaArrayDataSet(beta_array, transform)[source]¶ Torch Dataset just for beta matrix in numpy format.
Parameters: - beta_array : numpy.array
Beta value matrix in numpy form.
- transform :
Transforms data to torch tensor.
Attributes: - length :
Number CpGs.
- beta_array
- transform
-
class
methylnet.datasets.Transformer(convolutional=False, cpg_per_row=30000, l=None)[source]¶ Push methyl array sample to pytorch tensor, possibly turning into image.
Parameters: - convolutional : bool
Whether running convolutions on torch dataset.
- cpg_per_row : int
Number of CpGs per row of image if convolutional.
- l : tuple
Attributes that describe image.
Attributes: - shape : tuple
Shape of methylation array image for sample
- convolutional
- cpg_per_row
- l
Methods
generate(self)Generate function for transform.
-
methylnet.datasets.cat()¶ Concatenates the given sequence of
seqtensors in the given dimension. All tensors must either have the same shape (except in the concatenating dimension) or be empty.torch.cat()can be seen as an inverse operation fortorch.split()andtorch.chunk().torch.cat()can be best understood via examples.- Args:
- tensors (sequence of Tensors): any python sequence of tensors of the same type.
- Non-empty tensors provided must have the same shape, except in the cat dimension.
dim (int, optional): the dimension over which the tensors are concatenated out (Tensor, optional): the output tensor
Example:
>>> x = torch.randn(2, 3) >>> x tensor([[ 0.6580, -1.0969, -0.4614], [-0.1034, -0.5790, 0.1497]]) >>> torch.cat((x, x, x), 0) tensor([[ 0.6580, -1.0969, -0.4614], [-0.1034, -0.5790, 0.1497], [ 0.6580, -1.0969, -0.4614], [-0.1034, -0.5790, 0.1497], [ 0.6580, -1.0969, -0.4614], [-0.1034, -0.5790, 0.1497]]) >>> torch.cat((x, x, x), 1) tensor([[ 0.6580, -1.0969, -0.4614, 0.6580, -1.0969, -0.4614, 0.6580, -1.0969, -0.4614], [-0.1034, -0.5790, 0.1497, -0.1034, -0.5790, 0.1497, -0.1034, -0.5790, 0.1497]])
-
methylnet.datasets.get_methylation_dataset(methylation_array, outcome_col, convolutional=False, cpg_per_row=1200, predict=False, categorical=False, categorical_encoder=False, generate=False)[source]¶ Turn methylation array into pytorch dataset.
Parameters: - methylation_array : MethylationArray
Input MethylationArray.
- outcome_col : str
Pheno column to train on.
- convolutional : bool
Whether running CNN on methylation data
- cpg_per_row : int
If convolutional, number of cpgs per image row.
- predict : bool
Running prediction algorithm vs VAE.
- categorical : bool
Whether training on categorical vs continuous variables.
- categorical_encoder :
Scikit learn encoder.
Returns: - Pytorch Dataset
-
methylnet.datasets.stack()¶ Concatenates sequence of tensors along a new dimension.
All tensors need to be of the same size.
- Arguments:
seq (sequence of Tensors): sequence of tensors to concatenate dim (int): dimension to insert. Has to be between 0 and the number
of dimensions of concatenated tensors (inclusive)out (Tensor, optional): the output tensor
hyperparameter_scans.py¶
Run randomized grid search to find ideal model hyperparameters, with possible deployments to batch system for scalability.
-
methylnet.hyperparameter_scans.coarse_scan(hyperparameter_input_csv, hyperparameter_output_log, generate_input, job_chunk_size, stratify_column, reset_all, torque, gpu, gpu_node, nohup, mlp=False, custom_jobs=[], model_complexity_factor=0.9, set_beta=-1.0, n_jobs=4, categorical=True, add_softmax=False, additional_command='', cuda=True, new_grid={})[source]¶ Perform randomized hyperparameter grid search
Parameters: - hyperparameter_input_csv : type
CSV file containing hyperparameter inputs.
- hyperparameter_output_log : type
CSV file containing prior runs.
- generate_input : type
Generate hyperparameter input csv.
- job_chunk_size : type
Number of jobs to be launched at same time.
- stratify_column : type
Performing classification?
- reset_all : type
Rerun all jobs previously scanned.
- torque : type
Run jobs using torque.
- gpu : type
What GPU to use, set to -1 to be agnostic to GPU selection.
- gpu_node : type
What GPU to use, set to -1 to be agnostic to GPU selection, for torque submission.
- nohup : type
Launch jobs using nohup.
- mlp : type
If running prediction job (classification/regression) after VAE.
- custom_jobs : type
Supply custom job parameters to be run.
- model_complexity_factor : type
Degree of neural network model complexity for hyperparameter search. Search for less wide networks with a lower complexity value, bounded between 0 and infinity.
- set_beta : type
Don’t hyperparameter scan over beta (KL divergence weight), and set it to value.
- n_jobs : type
Number of jobs to generate.
- categorical : type
Classification task?
- add_softmax : type
Add softmax layer at end of neural network.
- cuda : type
Whether to use GPU?
-
methylnet.hyperparameter_scans.find_top_jobs(hyperparameter_input_csv, hyperparameter_output_log, n_top_jobs, crossover_p=0, val_loss_column='min_val_loss')[source]¶ Finds top performing jobs from hyper parameter scan to rerun and cross-over parameters.
Parameters: - hyperparameter_input_csv : str
CSV file containing hyperparameter inputs.
- hyperparameter_output_log : str
CSV file containing prior runs.
- n_top_jobs : int
Number of top jobs to select
- crossover_p : float
Rate of cross over of reused hyperparameters
- val_loss_column : str
Loss column used to select top jobs
Returns: - list
List of list of new parameters for jobs to run.
-
methylnet.hyperparameter_scans.generate_topology(topology_grid, probability_decay_factor=0.9)[source]¶ Generates list denoting neural network topology, list of hidden layer sizes.
Parameters: - topology_grid : list
List of different hidden layer sizes (number neurons) to choose from.
- probability_decay_factor : float
Degree of neural network model complexity for hyperparameter search. Search for less wide networks with a lower complexity value, bounded between 0 and infinity.
Returns: - list
List of hidden layer sizes.
torque_jobs.py¶
Wraps and runs your commands through torque.
-
methylnet.torque_jobs.assemble_replace_dict(command, use_gpu, additions, queue, time, ngpu)[source]¶ Create dictionary to update BASH submission script for torque.
Parameters: - command : type
Command to executer through torque.
- use_gpu : type
GPUs needed?
- additions : type
Additional commands to add (eg. module loads).
- queue : type
Queue to place job in.
- time : type
How many hours to run job for.
- ngpu : type
Number of GPU to use.
Returns: - Dict
Dictionary used to update Torque Script.
-
methylnet.torque_jobs.assemble_run_torque(command, use_gpu, additions, queue, time, ngpu, additional_options='')[source]¶ Runs torque job after passing commands to setup bash file.
Parameters: - command : type
Command to executer through torque.
- use_gpu : type
GPUs needed?
- additions : type
Additional commands to add (eg. module loads).
- queue : type
Queue to place job in.
- time : type
How many hours to run job for.
- ngpu : type
Number of GPU to use.
- additional_options : type
Additional options to pass to Torque scheduler.
Returns: - job
Custom job name.
-
methylnet.torque_jobs.run_torque_job_(replace_dict, additional_options='')[source]¶ Run torque job after creating submission script.
Parameters: - replace_dict : type
Dictionary used to replace information in bash script to run torque job.
- additional_options : type
Additional options to pass scheduler.
Returns: - str
Custom torque job name.
interpretation_classes.py¶
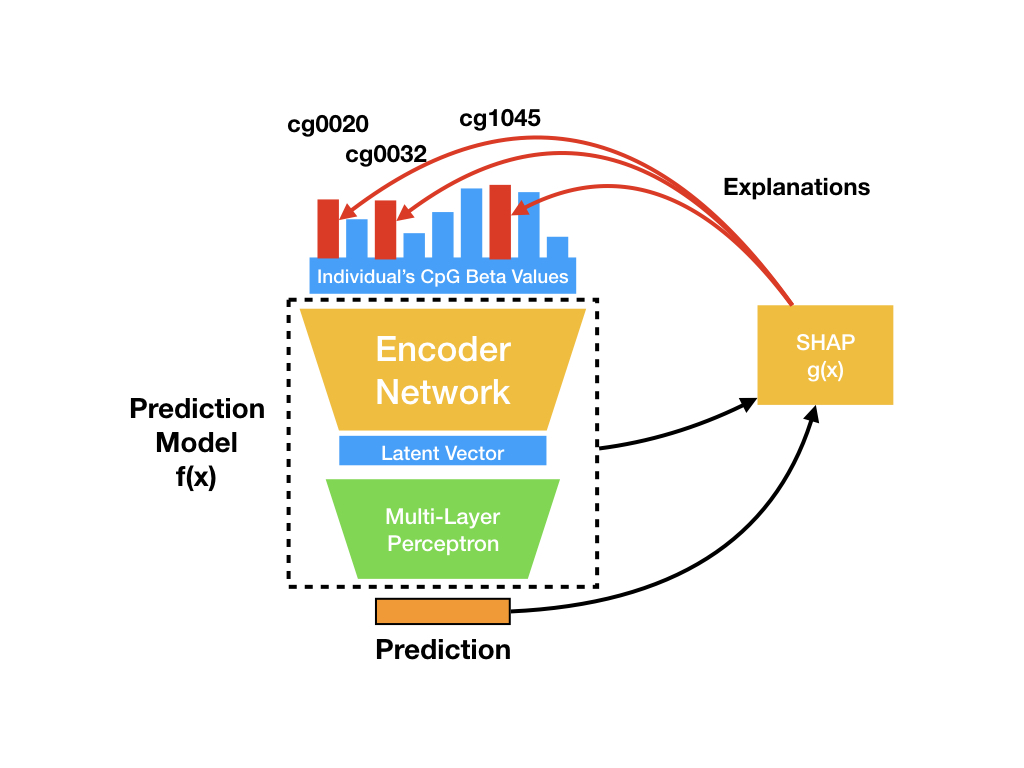
Contains core classes and functions to extracting explanations for the predictions at single samples, and then interrogates important CpGs for biological plausibility.
-
class
methylnet.interpretation_classes.BioInterpreter(dict_top_cpgs)[source]¶ Interrogate CpGs found to be important from SHAP through enrichment and overlap tests.
Parameters: - dict_top_cpgs : type
Dictionary of topCpGs output from ShapleyExplorer object
Attributes: - top_cpgs : type
Dictionary of top cpgs to be interrogated.
Methods
get_nearby_cpg_shapleys(self, all_cpgs, max_gap)Query for CpGs that are within a max_gap of the topCpGs in the genome, helps find cpgs that could be highly correlated and thus also important. gometh(self[, collection, allcpgs, …])Run GO, KEGG, GSEA analyses or return nearby genes. return_overlap_score(self[, set_cpgs, …])Perform overlap test, overlapping top CpGs with IDOL CpGs, age related cpgs, etc. run_lola(self[, all_cpgs, lola_db, cores, …])Run LOLA enrichment test. -
get_nearby_cpg_shapleys(self, all_cpgs, max_gap)[source]¶ Query for CpGs that are within a max_gap of the topCpGs in the genome, helps find cpgs that could be highly correlated and thus also important.
Parameters: - all_cpgs : type
List of all cpgs in methylationarray.
- max_gap : type
Max radius to search for nearby CpGs.
Returns: - Dict
Results for each class/indiv’s top CpGs, in the form of nearby CpGs to each of these CpGs sets.
-
gometh(self, collection='GO', allcpgs=[], length_output=20, gsea_analyses=[], gsea_pickle='')[source]¶ Run GO, KEGG, GSEA analyses or return nearby genes.
Parameters: - collection : type
GO, KEGG, GSEA, GENE overlaps?
- allcpgs : type
All CpGs defines CpG universe, empty if use all CpGs, smaller group of CpGs may yield more significant results.
- length_output : type
How many lines should the output be, maximum number of results to print.
- gsea_analyses : type
GSEA analyses/collections to target.
- gsea_pickle : type
Location of gsea pickle containing gene sets, may need to run download_help_data to acquire.
Returns: - Dict
Dictionary containing results from each test run on all of the keys in top_cpgs.
-
return_overlap_score(self, set_cpgs='IDOL', platform='450k', all_cpgs=[], output_csv='output_bio_intersect.csv', extract_library=False)[source]¶ Perform overlap test, overlapping top CpGs with IDOL CpGs, age related cpgs, etc.
Parameters: - set_cpgs : type
Set of reference cpgs.
- platform : type
450K or 850K
- all_cpgs : type
All CpGs to build a universe.
- output_csv : type
Output CSV for results of overlaps between classes, how much doo they share these CpGs overlapped.
- extract_library : type
Can extract a list of the CpGs that ended up beinng overlapped with.
Returns: - List
Optional output of overlapped library of CpGs.
-
run_lola(self, all_cpgs=[], lola_db='', cores=8, collections=[], depletion=False)[source]¶ Run LOLA enrichment test.
Parameters: - all_cpgs : type
CpG universe for LOLA.
- lola_db : type
Location of LOLA database, can be downloaded using methylnet-interpret. Set to extended or core, and collections correspond to these.
- cores : type
Number of cores to use.
- collections : type
LOLA collections to run, leave empty to run all.
- depletion : type
Set true to look for depleted regions over enriched.
Returns: - type
Description of returned object.
-
class
methylnet.interpretation_classes.CpGExplainer(prediction_function=None, cuda=False)[source]¶ Produces SHAPley explanation scores for individual predictions, approximating a complex model with a basic linear one, one coefficient per CpG per individual.
Parameters: - prediction_function : type
Model or function that makes predictions on the data, can be sklearn .predict method or pytorch forward pass, etc…
- cuda : type
Run on GPUs?
Attributes: - explainer : type
Type of SHAPley explanation method; kernel (uses LIME model agnostic explainer), deep (uses DeepLift) or Gradient (integrated gradients and backprop for explanations)?
- prediction_function
- cuda
Methods
build_explainer(self, train_methyl_array[, …])Builds SHAP explainer using background samples. classifier_assign_scores_to_shap_data(self, …)Assigns the SHAP scores to a SHAPley data object, populating nested dictionaries (containing class-individual information) with SHAPley information and “top CpGs”. feature_select(self, methyl_array, …)Perform feature selection based on the best overall SHAP scores across all samples. from_explainer(method, cuda)Load custom SHAPley explainer regressor_assign_scores_to_shap_data(self, …)Assigns the SHAP scores to a SHAPley data object, populating nested dictionaries (containing multioutput and single output regression-individual information) with SHAPley information and “top CpGs”. return_shapley_predictions(self, …[, encoder])Method in development or may be deprecated. return_shapley_scores(self, …[, …])Generate explanations for individual predictions, in the form of SHAPley scores for supplied test data. -
build_explainer(self, train_methyl_array, method='kernel', batch_size=100)[source]¶ Builds SHAP explainer using background samples.
Parameters: - train_methyl_array : type
Train Methylation Array from which to populate background samples to make estimated explanations.
- method : type
SHAP explanation method?
- batch_size : type
Break up prediction explanation creation into smaller batch sizes for lower memory consumption.
-
classifier_assign_scores_to_shap_data(self, test_methyl_array, n_top_features, interest_col='disease', prediction_classes=None)[source]¶ Assigns the SHAP scores to a SHAPley data object, populating nested dictionaries (containing class-individual information) with SHAPley information and “top CpGs”.
Parameters: - test_methyl_array : type
Testing MethylationArray.
- n_top_features : type
Number of top SHAP scores to use for a subdict called “top CpGs”
- interest_col : type
Column in pheno sheet from which class names reside.
- prediction_classes : type
User supplied prediction classes.
-
feature_select(self, methyl_array, n_top_features)[source]¶ Perform feature selection based on the best overall SHAP scores across all samples.
Parameters: - methyl_array : type
MethylationArray to run feature selection on.
- n_top_features : type
Number of CpGs to select.
Returns: - MethylationArray
Subsetted by top overall CpGs, overall positive contributions to prediction. May need to update.
-
regressor_assign_scores_to_shap_data(self, test_methyl_array, n_top_features, cell_names=[])[source]¶ Assigns the SHAP scores to a SHAPley data object, populating nested dictionaries (containing multioutput and single output regression-individual information) with SHAPley information and “top CpGs”.
Parameters: - test_methyl_array : type
Testing MethylationArray.
- n_top_features : type
Number of top SHAP scores to use for a subdict called “top CpGs”
- cell_names : type
If multi-output regression, create separate regression classes to access for all individuals.
-
return_shapley_predictions(self, test_methyl_array, sample_name, interest_col, encoder=None)[source]¶ Method in development or may be deprecated.
-
return_shapley_scores(self, test_methyl_array, n_samples, n_outputs, shap_sample_batch_size=None, top_outputs=None)[source]¶ Generate explanations for individual predictions, in the form of SHAPley scores for supplied test data. One SHAP score per individual prediction per CpG, and more if multiclass/output. For multiclass/multivariate outcomes, scores are generated for each outcome class.
Parameters: - test_methyl_array : type
Testing MethylationArray.
- n_samples : type
Number of SHAP score samples to produce. More SHAP samples provides convergence to correct score.
- n_outputs : type
Number of outcome classes.
- shap_sample_batch_size : type
If not None, break up SHAP score sampling into batches of this size to be averaged.
- top_outputs : type
For deep explainer, limit number of output classes due to memory consumption.
Returns: - type
Description of returned object.
-
class
methylnet.interpretation_classes.DistanceMatrixCompute(methyl_array, pheno_col)[source]¶ From any embeddings, calculate pairwise distances between classes that label embeddings.
Parameters: - methyl_array : MethylationArray
Methylation array storing beta and pheno data.
- pheno_col : str
Column name from which to extract sample names from and group by class.
Attributes: - col : pd.DataFrame
Column of pheno array.
- classes : np.array
Unique classes of pheno column.
- embeddings : pd.DataFrame
Embeddings of beta values.
Methods
calculate_p_values(self)Compute pairwise p-values between different clusters using manova. compute_distances(self[, metric, trim])Compute distance matrix between classes by average distances between points of classes. return_distances(self)Return the distance matrix return_p_values(self)Return the distance matrix -
calculate_p_values(self)[source]¶ Compute pairwise p-values between different clusters using manova.
-
compute_distances(self, metric='cosine', trim=0.0)[source]¶ Compute distance matrix between classes by average distances between points of classes.
Parameters: - metric : str
Scikit-learn distance metric.
-
class
methylnet.interpretation_classes.PlotCircos[source]¶ Plot Circos Diagram using ggbio (regular circos software may be better)
Attributes: - generate_base_plot : type
Function to generate base plot
- add_plot_data : type
Function to add more plot data
- generate_final_plot : type
Function to plot data using Circos
Methods
plot_cpgs(self, top_cpgs[, output_dir])Plot top CpGs location in genome.
-
class
methylnet.interpretation_classes.ShapleyData[source]¶ Store SHAP results that stores feature importances of CpGs on varying degrees granularity.
Attributes: - top_cpgs : type
Quick accessible CpGs that have the n highest SHAP scores.
- shapley_values : type
Storing all SHAPley values for CpGs for varying degrees granularity. For classification problems, this only includes SHAP scores particular to the actual class.
Methods
add_class(self, class_name, shap_df, cpgs, …)Store SHAP scores for particular class to Shapley_Data class. add_global_importance(self, …)Add overall feature importances, globally across all samples. from_pickle(input_pkl[, from_dict])Load SHAPley data from pickle. to_pickle(self, output_pkl[, output_dict])Export Shapley data to pickle. -
add_class(self, class_name, shap_df, cpgs, n_top_cpgs, add_top_negative=False, add_top_abs=False)[source]¶ Store SHAP scores for particular class to Shapley_Data class. Save feature importances on granular level, explanations for individual and aggregate predictions.
Parameters: - class_name : type
Particular class name to be stored in dictionary structure.
- shap_df : type
SHAP data to pull from.
- cpgs : type
All CpGs.
- n_top_cpgs : type
Number of top CpGs for saving n top CpGs.
- add_top_negative : type
Only consider top negative scores.
-
add_global_importance(self, global_importance_shaps, cpgs, n_top_cpgs)[source]¶ Add overall feature importances, globally across all samples.
Parameters: - global_importance_shaps : type
Overall SHAP values to be saved, aggregated across all samples.
- cpgs : type
All CpGs.
- n_top_cpgs : type
Number of top CpGs to rank and save as well.
-
class
methylnet.interpretation_classes.ShapleyDataExplorer(shapley_data)[source]¶ Datatype used to explore saved ShapleyData.
Parameters: - shapley_data : type
ShapleyData instance to be explored.
Attributes: - indiv2class : type
Maps individuals to their classes used for quick look-up.
- shapley_data
Methods
add_abs_value_classes(self)WIP. add_bin_continuous_classes(self)Bin continuous outcome variable and extract top positive and negative CpGs for each new class. extract_class(self, class_name[, …])Extract the top cpgs from a class extract_individual(self, individual[, …])Extract the top cpgs from an individual extract_methylation_array(self, methyl_arr)Subset MethylationArray Beta values by some top SHAP CpGs from classes or overall. jaccard_similarity_top_cpgs(self, class_names)Calculate Jaccard Similarity matrix between classes and individuals within those classes based on how they share sets of CpGs. limit_number_top_cpgs(self, n_top_cpgs)Reduce the number of top CpGs. list_classes(self)List classes in ShapleyData object. list_individuals(self[, return_list])List the individuals in the ShapleyData object. regenerate_individual_shap_values(self, …)Use original SHAP scores to make nested dictionary of top CpGs based on shapley score, can do this for ABS SHAP or Negative SHAP scores as well. return_binned_shapley_data(self, …[, …])Converts existing shap data based on continuous variable predictions into categorical variable. return_cpg_sets(self)Return top sets of CpGs from classes and individuals, and the complementary set. return_global_importance_cpgs(self)Return overall globally important CpGs. return_shapley_data_by_methylation_status(…)Return dictionary containing two SHAPley datasets, each split by low/high levels of methylation. return_top_cpgs(self[, classes, …])Given list of classes and individuals, export a dictionary containing data frames of top CpGs and their SHAP scores. view_methylation(self, individual, methyl_arr)Use MethylationArray to output top SHAP values for individual with methylation data appended to output data frame. make_shap_scores_abs -
add_bin_continuous_classes(self)[source]¶ Bin continuous outcome variable and extract top positive and negative CpGs for each new class.
-
extract_class(self, class_name, class_intersect=False, get_shap_values=False)[source]¶ Extract the top cpgs from a class
Parameters: - class_name : type
Class to extract from?
- class_intersect : type
Bool to extract from aggregation of SHAP values from individuals of current class, should have been done already.
Returns: - DataFrame
Cpgs and SHAP Values
-
extract_individual(self, individual, get_shap_values=False)[source]¶ Extract the top cpgs from an individual
Parameters: - individual : type
Individual to extract from?
Returns: - Tuple
Class name of individual, DataFrame of Cpgs and SHAP Values
-
extract_methylation_array(self, methyl_arr, classes_only=True, global_vals=False, n_extract=1000, class_name='')[source]¶ Subset MethylationArray Beta values by some top SHAP CpGs from classes or overall.
Parameters: - methyl_arr : type
MethylationArray.
- classes_only : type
Setting this to False will include the overall top CpGs per class and the top CpGs for individuals in that class.
- global_vals : type
Use top CpGs overall, across the entire dataset.
- n_extract : type
Number of top CpGs to subset.
- class_name : type
Which class to subset top CpGs from? Blank to use union of all top CpGs.
Returns: - MethylationArray
Stores methylation beta and pheno data, reduced here by queried CpGs.
-
jaccard_similarity_top_cpgs(self, class_names, individuals=False, overall=False, cooccurrence=False)[source]¶ Calculate Jaccard Similarity matrix between classes and individuals within those classes based on how they share sets of CpGs.
Parameters: - class_names : type
Classes to include.
- individuals : type
Individuals to include.
- overall : type
Whether to use overall class top CpGs versus aggregate.
- cooccurrence : type
Output cooccurence instead of jaccard.
Returns: - pd.DataFrame
Similarity matrix.
-
limit_number_top_cpgs(self, n_top_cpgs)[source]¶ Reduce the number of top CpGs.
Parameters: - n_top_cpgs : type
Number top CpGs to retain.
Returns: - ShapleyData
Returns shapley data with fewer top CpGs.
-
list_individuals(self, return_list=False)[source]¶ List the individuals in the ShapleyData object.
Parameters: - return_list : type
Return list of individual names rather than a dictionary of class:individual key-value pairs.
Returns: - List or Dictionary
Class: Individual dictionary or individuals are elements of list
-
regenerate_individual_shap_values(self, n_top_cpgs, abs_val=False, neg_val=False)[source]¶ Use original SHAP scores to make nested dictionary of top CpGs based on shapley score, can do this for ABS SHAP or Negative SHAP scores as well.
Parameters: - n_top_cpgs : type
Description of parameter n_top_cpgs.
- abs_val : type
Description of parameter abs_val.
- neg_val : type
Description of parameter neg_val.
Returns: - type
Description of returned object.
-
return_binned_shapley_data(self, original_class_name, outcome_col, add_top_negative=False)[source]¶ Converts existing shap data based on continuous variable predictions into categorical variable.
Parameters: - original_class_name : type
Regression results were split into ClassName_pos and ClassName_neg, what is the ClassName?
- outcome_col : type
Feed in from pheno sheet one the column to bin samples on.
- add_top_negative : type
Looking to include negative SHAPs?
Returns: - ShapleyData
With new class labels, built from regression results.
-
return_cpg_sets(self)[source]¶ Return top sets of CpGs from classes and individuals, and the complementary set. Returns dictionary of sets of CpGs and the union of all the other sets minus the current set.
Returns: - cpg_sets
Dictionary of individuals and classes; CpGs contained in set for particular individual/class
- cpg_exclusion_sets
Dictionary of individuals and classes; CpGs not contained in set for particular individual/class
-
return_shapley_data_by_methylation_status(self, methyl_array, threshold)[source]¶ Return dictionary containing two SHAPley datasets, each split by low/high levels of methylation. Todo: Define this using median methylation value vs 0.5.
Parameters: - methyl_array : type
MethylationArray instance.
Returns: - dictionary
Contains shapley data by methylation status.
-
return_top_cpgs(self, classes=[], individuals=[], class_intersect=False, cpg_exclusion_sets=None, cpg_sets=None)[source]¶ Given list of classes and individuals, export a dictionary containing data frames of top CpGs and their SHAP scores.
Parameters: - classes : type
Higher level classes to extract CpGs from, list of classes to extract top CpGs from.
- individuals : type
Individual samples to extract top CpGs from.
- class_intersect : type
Whether the top CpGs should be chosen by aggregating the remaining individual scores.
- cpg_exclusion_sets : type
Dict of sets of CpGs, where these ones contain CpGs not particular to particular class.
- cpg_sets : type
Contains top CpGs found for each class.
Returns: - Dict
Top CpGs accessed and returned for further processing.
-
view_methylation(self, individual, methyl_arr)[source]¶ Use MethylationArray to output top SHAP values for individual with methylation data appended to output data frame.
Parameters: - individual : type
Individual to query from ShapleyData
- methyl_arr : type
Input MethylationArray
Returns: - class_name
- individual
- top_shap_df
Top Shapley DataFrame with top cpgs for individual, methylation data appended.
-
methylnet.interpretation_classes.cooccurrence_fn(list1, list2)[source]¶ Cooccurence of elements between two lists.
Parameters: - list1
- list2
Returns: - float
Cooccurence between elements in list.
-
methylnet.interpretation_classes.jaccard_similarity(list1, list2)[source]¶ Jaccard score between two lists.
Parameters: - list1
- list2
Returns: - float
Jaccard similarity between elements in list.
-
methylnet.interpretation_classes.main_prediction_function(n_workers, batch_size, model, cuda)[source]¶ Combine dataloader and prediction function into final prediction function that takes in tensor data, for Kernel Explanations.
Parameters: - n_workers : type
Number of workers.
- batch_size : type
Batch size for input data.
- model : type
Model/predidction function.
- cuda : type
Running on GPUs?
Returns: - function
Final prediction function.
-
methylnet.interpretation_classes.plot_lola_output_(lola_csv, plot_output_dir, description_col, cell_types)[source]¶ Plots any LOLA output in the form of a forest plot.
Parameters: - lola_csv : type
CSV containing lola results.
- plot_output_dir : type
Plot output directory.
- description_col : type
Column that will label the points on the plot.
- cell_types : type
Column containing cell-types, colors plots and are rows to be compared.
-
methylnet.interpretation_classes.return_dataloader_construct(n_workers, batch_size)[source]¶ Decorator to build dataloader compatible with KernelExplainer for SHAP.
Parameters: - n_workers : type
Number CPU.
- batch_size : type
Batch size to load data into pytorch.
Returns: - DataLoader
Pytorch dataloader.
-
methylnet.interpretation_classes.return_predict_function(model, cuda)[source]¶ Decorator to build the supplied prediction function, important for kernel explanations.
Parameters: - model : type
Prediction function with predict method.
- cuda : type
Run using cuda.
Returns: - function
Predict function.
-
methylnet.interpretation_classes.return_shap_values(test_arr, explainer, method, n_samples, additional_opts)[source]¶ Return SHAP values, sampled a number of times, used in CpGExplainer class.
Parameters: - test_arr : type
Testing MethylationArray.
- explainer : type
SHAP explainer object.
- method : type
Method of explaining.
- n_samples : type
Number of samples to estimate SHAP scores.
- additional_opts : type
Additional options to be passed into non-kernel/gradient methods.
Returns: - np.array/list
Shapley scores.
models.py¶
Contains core PyTorch Models for running VAE and VAE-MLP.
-
class
methylnet.models.AutoEncoder(autoencoder_model, n_epochs, loss_fn, optimizer, cuda=True, kl_warm_up=0, beta=1.0, scheduler_opts={})[source]¶ Wraps Pytorch VAE module into Scikit-learn like interface for ease of training, validation and testing.
Parameters: - autoencoder_model : type
Pytorch VAE Model to supply.
- n_epochs : type
Number of epochs to train for.
- loss_fn : type
Pytorch loss function for reconstruction error.
- optimizer : type
Pytorch Optimizer.
- cuda : type
GPU?
- kl_warm_up : type
Number of epochs until fully utilizing KLLoss, begin saving models here.
- beta : type
Weighting for KLLoss.
- scheduler_opts : type
Options to feed learning rate scheduler, which modulates learning rate of optimizer.
Attributes: - model : type
Pytorch VAE model.
- scheduler : type
Learning rate scheduler object.
- vae_animation_fname : type
Save VAE embeddings evolving over epochs to this file name. Defunct for now.
- loss_plt_fname : type
Where to save loss curves. This has been superceded by plot_training_curves in methylnet-visualize command.
- plot_interval : type
How often to plot data; defunct.
- embed_interval : type
How often to embed; defunct.
- validation_set : type
MethylationArray DataLoader, produced from Pytorch MethylationDataset of Validation MethylationArray.
- n_epochs
- loss_fn
- optimizer
- cuda
- kl_warm_up
- beta
Methods
add_validation_set(self, validation_data)Add validation data in the form of Validation DataLoader. fit(self, train_data)Fit VAE model to training data, best model returned with lowest validation loss over epochs. fit_transform(self, train_data)Fit VAE model and transform Methylation Array using VAE model. transform(self, train_data)Parameters: -
add_validation_set(self, validation_data)[source]¶ Add validation data in the form of Validation DataLoader. Adding this will use validation data for early termination / generalization of model to unseen data.
Parameters: - validation_data : type
Pytorch DataLoader housing validation MethylationDataset.
-
fit(self, train_data)[source]¶ Fit VAE model to training data, best model returned with lowest validation loss over epochs.
Parameters: - train_data : DataLoader
Training DataLoader that is loading MethylationDataset in batches.
Returns: - self
Autoencoder object with updated VAE model.
-
fit_transform(self, train_data)[source]¶ Fit VAE model and transform Methylation Array using VAE model.
Parameters: - train_data : type
Pytorch DataLoader housing training MethylationDataset.
Returns: - np.array
Latent Embeddings.
- np.array
Sample names from MethylationArray
- np.array
Outcomes from column of methylarray.
-
class
methylnet.models.MLPFinetuneVAE(mlp_model, n_epochs=None, loss_fn=None, optimizer_vae=None, optimizer_mlp=None, cuda=True, categorical=False, scheduler_opts={}, output_latent=True, train_decoder=False)[source]¶ Wraps VAE_MLP pytorch module into scikit-learn interface with fit, predict and fit_predict methods for ease-of-use model training/evaluation.
Parameters: - mlp_model : type
VAE_MLP model.
- n_epochs : type
Number epochs train for.
- loss_fn : type
Loss function, pytorch, CrossEntropy, BCE, MSE depending on outcome.
- optimizer_vae : type
Optimizer for VAE layers for finetuning original pretrained network.
- optimizer_mlp : type
Optimizer for new appended MLP layers.
- cuda : type
GPU?
- categorical : type
Classification or regression outcome?
- scheduler_opts : type
Options for learning rate scheduler, modulates learning rates for VAE and MLP.
- output_latent : type
Whether to output latent embeddings during evaluation.
- train_decoder : type
Retrain decoder to adjust for finetuning of VAE?
Attributes: - model : type
VAE_MLP.
- scheduler_vae : type
Learning rate modulator for VAE optimizer.
- scheduler_mlp : type
Learning rate modulator for MLP optimizer.
- loss_plt_fname : type
File where to plot loss over time; defunct.
- embed_interval : type
How often to return embeddings; defunct.
- validation_set : type
Validation set used for hyperparameter tuning and early stopping criteria for generalization.
- return_latent : type
Return embedding during evaluation?
- n_epochs
- loss_fn
- optimizer_vae
- optimizer_mlp
- cuda
- categorical
- output_latent
- train_decoder
Methods
add_validation_set(self, validation_data)Add validation data to reduce overfitting. fit(self, train_data)Fit MLP to training data to make predictions. predict(self, test_data)Short summary. -
add_validation_set(self, validation_data)[source]¶ Add validation data to reduce overfitting.
Parameters: - validation_data : type
Validation Dataloader MethylationDataset.
-
class
methylnet.models.TybaltTitusVAE(n_input, n_latent, hidden_layer_encoder_topology=[100, 100, 100], cuda=False)[source]¶ Pytorch NN Module housing VAE with fully connected layers and customizable topology.
Parameters: - n_input : type
Number of input CpGs.
- n_latent : type
Size of latent embeddings.
- hidden_layer_encoder_topology : type
List, length of list contains number of hidden layers for encoder, and each element is number of neurons, mirrored for decoder.
- cuda : type
GPU?
Attributes: - cuda_on : type
GPU?
- pre_latent_topology : type
Hidden layer topology for encoder.
- post_latent_topology : type
Mirrored hidden layer topology for decoder.
- encoder_layers : list
Encoder pytorch layers.
- encoder : type
Encoder layers wrapped into pytorch module.
- z_mean : type
Linear layer from last encoder layer to mean layer.
- z_var : type
Linear layer from last encoder layer to var layer.
- z_develop : type
Linear layer connecting sampled latent embedding to first layer decoder.
- decoder_layers : type
Decoder layers wrapped into pytorch module.
- output_layer : type
Linear layer connecting last decoder layer to output layer, which is same size as input..
- decoder : type
Wraps decoder_layers and output_layers into Sequential module.
- n_input
- n_latent
Methods
__call__(self, \*input, \*\*kwargs)Call self as a function. add_module(self, name, module)Adds a child module to the current module. apply(self, fn)Applies fnrecursively to every submodule (as returned by.children()) as well as self.buffers(self[, recurse])Returns an iterator over module buffers. children(self)Returns an iterator over immediate children modules. cpu(self)Moves all model parameters and buffers to the CPU. cuda(self[, device])Moves all model parameters and buffers to the GPU. decode(self, z)Decode latent embeddings back into reconstructed input. double(self)Casts all floating point parameters and buffers to doubledatatype.encode(self, x)Encode input into latent representation. eval(self)Sets the module in evaluation mode. extra_repr(self)Set the extra representation of the module float(self)Casts all floating point parameters and buffers to float datatype. forward(self, x)Return reconstructed output, mean and variance of embeddings. forward_predict(self, x)Forward pass from input to reconstructed input. get_latent_z(self, x)Encode X into reparameterized latent representation. half(self)Casts all floating point parameters and buffers to halfdatatype.load_state_dict(self, state_dict[, strict])Copies parameters and buffers from state_dictinto this module and its descendants.modules(self)Returns an iterator over all modules in the network. named_buffers(self[, prefix, recurse])Returns an iterator over module buffers, yielding both the name of the buffer as well as the buffer itself. named_children(self)Returns an iterator over immediate children modules, yielding both the name of the module as well as the module itself. named_modules(self[, memo, prefix])Returns an iterator over all modules in the network, yielding both the name of the module as well as the module itself. named_parameters(self[, prefix, recurse])Returns an iterator over module parameters, yielding both the name of the parameter as well as the parameter itself. parameters(self[, recurse])Returns an iterator over module parameters. register_backward_hook(self, hook)Registers a backward hook on the module. register_buffer(self, name, tensor)Adds a persistent buffer to the module. register_forward_hook(self, hook)Registers a forward hook on the module. register_forward_pre_hook(self, hook)Registers a forward pre-hook on the module. register_parameter(self, name, param)Adds a parameter to the module. sample_z(self, mean, logvar)Sample latent embeddings, reparameterize by adding noise to embedding. state_dict(self[, destination, prefix, …])Returns a dictionary containing a whole state of the module. to(self, \*args, \*\*kwargs)Moves and/or casts the parameters and buffers. train(self[, mode])Sets the module in training mode. type(self, dst_type)Casts all parameters and buffers to dst_type.zero_grad(self)Sets gradients of all model parameters to zero. share_memory -
decode(self, z)[source]¶ Decode latent embeddings back into reconstructed input.
Parameters: - z : type
Reparameterized latent embedding.
Returns: - torch.tensor
Reconstructed input.
-
encode(self, x)[source]¶ Encode input into latent representation.
Parameters: - x : type
Input methylation data.
Returns: - torch.tensor
Learned mean vector of embeddings.
- torch.tensor
Learned variance of learned mean embeddings.
-
class
methylnet.models.VAE_MLP(vae_model, n_output, categorical=False, hidden_layer_topology=[100, 100, 100], dropout_p=0.2, add_softmax=False)[source]¶ VAE_MLP, pytorch module used to both finetune VAE embeddings and simultaneously train downstream MLP layers for classification/regression tasks.
Parameters: - vae_model : type
VAE pytorch model for methylation data.
- n_output : type
Number of outputs at end of model.
- categorical : type
Classification or regression problem?
- hidden_layer_topology : type
Hidden Layer topology, list of size number of hidden layers for MLP and each element contains number of neurons per layer.
- dropout_p : type
Apply dropout regularization to reduce overfitting.
- add_softmax : type
Softmax the output before evaluation.
Attributes: - vae : type
Pytorch VAE module.
- topology : type
List with hidden layer topology of MLP.
- mlp_layers : type
All MLP layers (# layers and neurons per layer)
- output_layer : type
nn.Linear connecting last MLP layer and output nodes.
- mlp : type
nn.Sequential wraps all layers into sequential ordered pytorch module.
- output_z : type
Whether to output latent embeddings.
- n_output
- categorical
- add_softmax
- dropout_p
Methods
__call__(self, \*input, \*\*kwargs)Call self as a function. add_module(self, name, module)Adds a child module to the current module. apply(self, fn)Applies fnrecursively to every submodule (as returned by.children()) as well as self.buffers(self[, recurse])Returns an iterator over module buffers. children(self)Returns an iterator over immediate children modules. cpu(self)Moves all model parameters and buffers to the CPU. cuda(self[, device])Moves all model parameters and buffers to the GPU. decode(self, z)Run VAE decoder on embeddings. double(self)Casts all floating point parameters and buffers to doubledatatype.eval(self)Sets the module in evaluation mode. extra_repr(self)Set the extra representation of the module float(self)Casts all floating point parameters and buffers to float datatype. forward(self, x)Pass data in to return predictions and embeddings. forward_embed(self, x)Return predictions, latent embeddings and reconstructed input. forward_predict(self, x)Make predictions, based on output_z, either output predictions or output embeddings. half(self)Casts all floating point parameters and buffers to halfdatatype.load_state_dict(self, state_dict[, strict])Copies parameters and buffers from state_dictinto this module and its descendants.modules(self)Returns an iterator over all modules in the network. named_buffers(self[, prefix, recurse])Returns an iterator over module buffers, yielding both the name of the buffer as well as the buffer itself. named_children(self)Returns an iterator over immediate children modules, yielding both the name of the module as well as the module itself. named_modules(self[, memo, prefix])Returns an iterator over all modules in the network, yielding both the name of the module as well as the module itself. named_parameters(self[, prefix, recurse])Returns an iterator over module parameters, yielding both the name of the parameter as well as the parameter itself. parameters(self[, recurse])Returns an iterator over module parameters. register_backward_hook(self, hook)Registers a backward hook on the module. register_buffer(self, name, tensor)Adds a persistent buffer to the module. register_forward_hook(self, hook)Registers a forward hook on the module. register_forward_pre_hook(self, hook)Registers a forward pre-hook on the module. register_parameter(self, name, param)Adds a parameter to the module. state_dict(self[, destination, prefix, …])Returns a dictionary containing a whole state of the module. to(self, \*args, \*\*kwargs)Moves and/or casts the parameters and buffers. toggle_latent_z(self)Toggle whether to output latent embeddings during forward pass. train(self[, mode])Sets the module in training mode. type(self, dst_type)Casts all parameters and buffers to dst_type.zero_grad(self)Sets gradients of all model parameters to zero. share_memory -
decode(self, z)[source]¶ Run VAE decoder on embeddings.
Parameters: - z : type
Embeddings.
Returns: - torch.tensor
Reconstructed Input.
-
forward(self, x)[source]¶ Pass data in to return predictions and embeddings.
Parameters: - x : type
Input data.
Returns: - torch.tensor
Predictions
- torch.tensor
Embeddings
-
forward_embed(self, x)[source]¶ Return predictions, latent embeddings and reconstructed input.
Parameters: - x : type
Input data
Returns: - torch.tensor
Predictions
- torch.tensor
Embeddings
- torch.tensor
Reconstructed input.
-
methylnet.models.project_vae(model, loader, cuda=True)[source]¶ Return Latent Embeddings of any data supplied to it.
Parameters: - model : type
VAE Pytorch Model.
- loader : type
Loads data one batch at a time.
- cuda : type
GPU?
Returns: - np.array
Latent Embeddings.
- np.array
Sample names from MethylationArray
- np.array
Outcomes from column of methylarray.
-
methylnet.models.test_mlp(model, loader, categorical, cuda=True, output_latent=True)[source]¶ Evaluate MLP on testing set, output predictions.
Parameters: - model : type
VAE_MLP model.
- loader : type
DataLoader with MethylationDataSet
- categorical : type
Categorical or continuous predictions.
- cuda : type
GPU?
- output_latent : type
Output latent embeddings in addition to predictions?
Returns: - np.array
Predictions
- np.array
Ground truth
- np.array
Latent Embeddings
- np.array
Sample names.
-
methylnet.models.train_decoder_(model, x, z)[source]¶ Run if retraining decoder to adjust for adjusted latent embeddings during finetuning of embedding layers for VAE_MLP.
Parameters: - model : type
VAE_MLP model.
- x : type
Input methylation data.
- z : type
Latent Embeddings
Returns: - nn.Module
VAE_MLP module with updated decoder parameters.
- float
Reconstruction loss over all batches.
-
methylnet.models.train_mlp(model, loader, loss_func, optimizer_vae, optimizer_mlp, cuda=True, categorical=False, train_decoder=False)[source]¶ Train Multi-layer perceptron appended to latent embeddings of VAE via transfer learning. Do this for one iteration.
Parameters: - model : type
VAE_MLP model.
- loader : type
DataLoader with MethylationDataset.
- loss_func : type
Loss function (BCE, CrossEntropy, MSE).
- optimizer_vae : type
Optimizer for pytorch VAE.
- optimizer_mlp : type
Optimizer for outcome MLP layers.
- cuda : type
GPU?
- categorical : type
Predicting categorical or continuous outcomes.
- train_decoder : type
Retrain decoder during training loop to adjust for fine-tuned embeddings.
Returns: - nn.Module
Training VAE_MLP model with updated parameters.
- float
Training loss over all batches
-
methylnet.models.train_vae(model, loader, loss_func, optimizer, cuda=True, epoch=0, kl_warm_up=0, beta=1.0)[source]¶ Function for parameter update during VAE training for one iteration.
Parameters: - model : type
VAE torch model
- loader : type
Data loader, generator that calls batches of data.
- loss_func : type
Loss function for reconstruction error, nn.BCELoss or MSELoss
- optimizer : type
SGD or Adam pytorch optimizer.
- cuda : type
GPU?
- epoch : type
Epoch of training, passed in from outer loop.
- kl_warm_up : type
How many epochs until model is fully utilizes KL Loss.
- beta : type
Weight given to KL Loss.
Returns: - nn.Module
Pytorch VAE model
- float
Total Training Loss across all batches
- float
Total Training reconstruction loss across all batches
- float
Total KL Loss across all batches
-
methylnet.models.vae_loss(output, input, mean, logvar, loss_func, epoch, kl_warm_up=0, beta=1.0)[source]¶ Function to calculate VAE Loss, Reconstruction Loss + Beta KLLoss.
Parameters: - output : torch.tensor
Reconstructed output from autoencoder.
- input : torch.tensor
Original input data.
- mean : type
Learned mean tensor for each sample point.
- logvar : type
Variation around that mean sample point, learned from reparameterization.
- loss_func : type
Loss function for reconstruction loss, MSE or BCE.
- epoch : type
Epoch of training.
- kl_warm_up : type
Number of epochs until fully utilizing KLLoss, begin saving models here.
- beta : type
Weighting for KLLoss.
Returns: - torch.tensor
Total loss
- torch.tensor
Recon loss
- torch.tensor
KL loss
-
methylnet.models.val_decoder_(model, x, z)[source]¶ Validation Loss over decoder.
Parameters: - model : type
VAE_MLP model.
- x : type
Input methylation data.
- z : type
Latent Embeddings
Returns: - float
Reconstruction loss over all batches.
-
methylnet.models.val_mlp(model, loader, loss_func, cuda=True, categorical=False, train_decoder=False)[source]¶ Find validation loss of VAE_MLP over one Epoch.
Parameters: - model : type
VAE_MLP model.
- loader : type
DataLoader with MethylationDataset.
- loss_func : type
Loss function (BCE, CrossEntropy, MSE).
- cuda : type
GPU?
- categorical : type
Predicting categorical or continuous outcomes.
- train_decoder : type
Retrain decoder during training loop to adjust for fine-tuned embeddings.
Returns: - nn.Module
VAE_MLP model.
- float
Validation loss over all batches
-
methylnet.models.val_vae(model, loader, loss_func, optimizer, cuda=True, epoch=0, kl_warm_up=0, beta=1.0)[source]¶ Function for validation loss computation during VAE training for one epoch.
Parameters: - model : type
VAE torch model
- loader : type
Validation Data loader, generator that calls batches of data.
- loss_func : type
Loss function for reconstruction error, nn.BCELoss or MSELoss
- optimizer : type
SGD or Adam pytorch optimizer.
- cuda : type
GPU?
- epoch : type
Epoch of training, passed in from outer loop.
- kl_warm_up : type
How many epochs until model is fully utilizes KL Loss.
- beta : type
Weight given to KL Loss.
Returns: - nn.Module
Pytorch VAE model
- float
Total Validation Loss across all batches
- float
Total Validation reconstruction loss across all batches
- float
Total Validation KL Loss across all batches
plotter.py¶
Plotting mechanisms for training that are now defuct.
-
class
methylnet.plotter.Plot(title, xlab='vae1', ylab='vae2', data=[])[source]¶ Stores plotting information; defunct, superceded, see methylnet-visualize.
Methods
return_img
schedulers.py¶
Learning rate schedulers that help enable better and more generalizable models.
-
class
methylnet.schedulers.CosineAnnealingWithRestartsLR(optimizer, T_max, eta_min=0, last_epoch=-1, T_mult=1.0, alpha_decay=1.0)[source]¶ Borrowed from: https://github.com/mpyrozhok/adamwr/blob/master/cyclic_scheduler.py Needs to be updated to reflect newest changes. From original docstring: Set the learning rate of each parameter group using a cosine annealing schedule, where \(\eta_{max}\) is set to the initial lr and \(T_{cur}\) is the number of epochs since the last restart in SGDR:
\[\eta_t = \eta_{min} + \frac{1}{2}(\eta_{max} - \eta_{min})(1 + \cos(\frac{T_{cur}}{T_{max}}\pi))\]When last_epoch=-1, sets initial lr as lr. It has been proposed in
SGDR: Stochastic Gradient Descent with Warm Restarts. This implements the cosine annealing part of SGDR, the restarts and number of iterations multiplier.
- Args:
- optimizer (Optimizer): Wrapped optimizer. T_max (int): Maximum number of iterations. T_mult (float): Multiply T_max by this number after each restart. Default: 1. eta_min (float): Minimum learning rate. Default: 0. last_epoch (int): The index of last epoch. Default: -1.
Attributes: - step_n
Methods
load_state_dict(self, state_dict)Loads the schedulers state. state_dict(self)Returns the state of the scheduler as a dict.cosine get_lr restart step
-
class
methylnet.schedulers.Scheduler(optimizer=None, opts={'T_max': 10, 'T_mult': 2, 'eta_min': 5e-08, 'lr_scheduler_decay': 0.5, 'scheduler': 'null'})[source]¶ Scheduler class that modulates learning rate of torch optimizers over epochs.
Parameters: - optimizer : type
torch.Optimizer object
- opts : type
Options of setting the learning rate scheduler, see default.
Attributes: - schedulers : type
Different types of schedulers to choose from.
- scheduler_step_fn : type
How scheduler updates learning rate.
- initial_lr : type
Initial set learning rate.
- scheduler_choice : type
What scheduler type was chosen.
- scheduler : type
Scheduler object chosen that will more directly update optimizer LR.
Methods
get_lr(self)Return current learning rate. step(self)Update optimizer learning rate
methylnet-embed¶
methylnet-embed [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
launch_hyperparameter_scan¶
Launch randomized grid search of neural network hyperparameters.
methylnet-embed launch_hyperparameter_scan [OPTIONS]
Options
-
-hcsv,--hyperparameter_input_csv<hyperparameter_input_csv>¶ CSV file containing hyperparameter inputs. [default: embeddings/embed_hyperparameters_scan_input.csv]
-
-hl,--hyperparameter_output_log<hyperparameter_output_log>¶ CSV file containing prior runs. [default: embeddings/embed_hyperparameters_log.csv]
-
-g,--generate_input¶ Generate hyperparameter input csv.
-
-c,--job_chunk_size<job_chunk_size>¶ If not series, chunk up and run these number of commands at once..
-
-sc,--stratify_column<stratify_column>¶ Column to stratify samples on. [default: disease]
-
-r,--reset_all¶ Run all jobs again.
-
-t,--torque¶ Submit jobs on torque.
-
-gpu,--gpu<gpu>¶ If torque submit, which gpu to use. [default: -1]
-
-gn,--gpu_node<gpu_node>¶ If torque submit, which gpu node to use. [default: -1]
-
-nh,--nohup¶ Nohup launch jobs.
-
-mc,--model_complexity_factor<model_complexity_factor>¶ Degree of neural network model complexity for hyperparameter search. Search for less wide and less deep networks with a lower complexity value, bounded between 0 and infinity. [default: 1.0]
-
-b,--set_beta<set_beta>¶ Set beta value, bounded between 0 and infinity. Set to -1 [default: 1.0]
-
-j,--n_jobs<n_jobs>¶ Number of jobs to generate.
-
-n,--n_jobs_relaunch<n_jobs_relaunch>¶ Relaunch n top jobs from previous run. [default: 0]
-
-cp,--crossover_p<crossover_p>¶ Rate of crossover between hyperparameters. [default: 0.0]
-
-v,--val_loss_column<val_loss_column>¶ Validation loss column.
-
-a,--additional_command<additional_command>¶ Additional command to input for torque run.
-
-cu,--cuda¶ Use GPUs.
-
-grid,--hyperparameter_yaml<hyperparameter_yaml>¶ YAML file with custom subset of hyperparameter grid.
perform_embedding¶
Perform variational autoencoding on methylation dataset.
methylnet-embed perform_embedding [OPTIONS]
Options
-
-i,--train_pkl<train_pkl>¶ Input database for beta and phenotype data. [default: ./train_val_test_sets/train_methyl_array.pkl]
-
-o,--output_dir<output_dir>¶ Output directory for embeddings. [default: ./embeddings/]
-
-c,--cuda¶ Use GPUs.
-
-n,--n_latent<n_latent>¶ Number of latent dimensions. [default: 64]
-
-lr,--learning_rate<learning_rate>¶ Learning rate. [default: 0.001]
-
-wd,--weight_decay<weight_decay>¶ Weight decay of adam optimizer. [default: 0.0001]
-
-e,--n_epochs<n_epochs>¶ Number of epochs to train over. [default: 50]
-
-hlt,--hidden_layer_encoder_topology<hidden_layer_encoder_topology>¶ Topology of hidden layers, comma delimited, leave empty for one layer encoder, eg. 100,100 is example of 5-hidden layer topology. [default: ]
-
-kl,--kl_warm_up<kl_warm_up>¶ Number of epochs before introducing kl_loss. [default: 0]
-
-b,--beta<beta>¶ Weighting of kl divergence. [default: 1.0]
-
-s,--scheduler<scheduler>¶ Type of learning rate scheduler. [default: null]
-
-d,--decay<decay>¶ Learning rate scheduler decay for exp selection. [default: 0.5]
-
-t,--t_max<t_max>¶ Number of epochs before cosine learning rate restart. [default: 10]
-
-eta,--eta_min<eta_min>¶ Minimum cosine LR. [default: 1e-06]
-
-m,--t_mult<t_mult>¶ Multiply current restart period times this number given number of restarts. [default: 2.0]
-
-bce,--bce_loss¶ Use bce loss instead of MSE.
-
-bs,--batch_size<batch_size>¶ Batch size. [default: 50]
-
-vp,--val_pkl<val_pkl>¶ Validation Set Methylation Array Location. [default: ./train_val_test_sets/val_methyl_array.pkl]
-
-w,--n_workers<n_workers>¶ Number of workers. [default: 9]
-
-conv,--convolutional¶ Use convolutional VAE.
-
-hs,--height_kernel_sizes<height_kernel_sizes>¶ Heights of convolutional kernels.
-
-ws,--width_kernel_sizes<width_kernel_sizes>¶ Widths of convolutional kernels.
-
-v,--add_validation_set¶ Evaluate validation set.
-
-l,--loss_reduction<loss_reduction>¶ Type of reduction on loss function. [default: sum]
-
-hl,--hyperparameter_log<hyperparameter_log>¶ CSV file containing prior runs. [default: embeddings/embed_hyperparameters_log.csv]
-
-sc,--stratify_column<stratify_column>¶ Column to stratify samples on. [default: disease]
-
-j,--job_name<job_name>¶ Embedding job name. [default: embed_job]
methylnet-predict¶
methylnet-predict [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
classification_report¶
Generate classification report that gives results from classification tasks.
methylnet-predict classification_report [OPTIONS]
Options
-
-r,--results_pickle<results_pickle>¶ Results from training, validation, and testing. [default: predictions/results.p]
-
-o,--output_dir<output_dir>¶ Output directory. [default: results/]
-
-e,--categorical_encoder<categorical_encoder>¶ One hot encoder if categorical model. If path exists, then return top positive controbutions per samples of that class. Encoded values must be of sample class as interest_col. [default: ./predictions/one_hot_encoder.p]
-
-a,--average_mechanism<average_mechanism>¶ Output directory. [default: micro]
launch_hyperparameter_scan¶
Run randomized hyperparameter scan of neural network hyperparameters.
methylnet-predict launch_hyperparameter_scan [OPTIONS]
Options
-
-hcsv,--hyperparameter_input_csv<hyperparameter_input_csv>¶ CSV file containing hyperparameter inputs. [default: predictions/predict_hyperparameters_scan_input.csv]
-
-hl,--hyperparameter_output_log<hyperparameter_output_log>¶ CSV file containing prior runs. [default: predictions/predict_hyperparameters_log.csv]
-
-g,--generate_input¶ Generate hyperparameter input csv.
-
-c,--job_chunk_size<job_chunk_size>¶ If not series, chunk up and run these number of commands at once..
-
-ic,--interest_cols<interest_cols>¶ Column to stratify samples on.
-
-cat,--categorical¶ Whether to run categorical analysis or not.
-
-r,--reset_all¶ Run all jobs again.
-
-t,--torque¶ Submit jobs on torque.
-
-gpu,--gpu<gpu>¶ If torque submit, which gpu to use. [default: -1]
-
-gn,--gpu_node<gpu_node>¶ If torque submit, which gpu node to use. [default: -1]
-
-nh,--nohup¶ Nohup launch jobs.
-
-n,--n_jobs_relaunch<n_jobs_relaunch>¶ Relaunch n top jobs from previous run. [default: 0]
-
-cp,--crossover_p<crossover_p>¶ Rate of crossover between hyperparameters. [default: 0.0]
-
-mc,--model_complexity_factor<model_complexity_factor>¶ Degree of neural network model complexity for hyperparameter search. Search for less wide networks with a lower complexity value, bounded between 0 and infinity. [default: 1.0]
-
-j,--n_jobs<n_jobs>¶ Number of jobs to generate.
-
-v,--val_loss_column<val_loss_column>¶ Validation loss column.
-
-sft,--add_softmax¶ Add softmax for predicting probability distributions.
-
-a,--additional_command<additional_command>¶ Additional command to input for torque run.
-
-cu,--cuda¶ Use GPUs.
-
-grid,--hyperparameter_yaml<hyperparameter_yaml>¶ YAML file with custom subset of hyperparameter grid.
make_new_predictions¶
Run prediction model again to further assess outcome. Only evaluate prediction model.
methylnet-predict make_new_predictions [OPTIONS]
Options
-
-tp,--test_pkl<test_pkl>¶ Test database for beta and phenotype data. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-m,--model_pickle<model_pickle>¶ Pytorch model containing forward_predict method. [default: ./predictions/output_model.p]
-
-bs,--batch_size<batch_size>¶ Batch size. [default: 50]
-
-w,--n_workers<n_workers>¶ Number of workers. [default: 9]
-
-ic,--interest_cols<interest_cols>¶ Specify columns looking to make predictions on. [default: disease]
-
-cat,--categorical¶ Multi-class prediction. [default: False]
-
-c,--cuda¶ Use GPUs.
-
-e,--categorical_encoder<categorical_encoder>¶ One hot encoder if categorical model. If path exists, then return top positive controbutions per samples of that class. Encoded values must be of sample class as interest_col. [default: ./predictions/one_hot_encoder.p]
-
-o,--output_dir<output_dir>¶ Output directory for predictions. [default: ./new_predictions/]
make_prediction¶
Train prediction model by fine-tuning VAE and appending/training MLP to make classification/regression predictions on MethylationArrays.
methylnet-predict make_prediction [OPTIONS]
Options
-
-i,--train_pkl<train_pkl>¶ Input database for beta and phenotype data. [default: ./train_val_test_sets/train_methyl_array.pkl]
-
-tp,--test_pkl<test_pkl>¶ Test database for beta and phenotype data. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-vae,--input_vae_pkl<input_vae_pkl>¶ Trained VAE. [default: ./embeddings/output_model.p]
-
-o,--output_dir<output_dir>¶ Output directory for predictions. [default: ./predictions/]
-
-c,--cuda¶ Use GPUs.
-
-ic,--interest_cols<interest_cols>¶ Specify columns looking to make predictions on. [default: disease]
-
-cat,--categorical¶ Multi-class prediction. [default: False]
-
-do,--disease_only¶ Only look at disease, or text before subtype_delimiter.
-
-hlt,--hidden_layer_topology<hidden_layer_topology>¶ Topology of hidden layers, comma delimited, leave empty for one layer encoder, eg. 100,100 is example of 5-hidden layer topology. [default: ]
-
-lr_vae,--learning_rate_vae<learning_rate_vae>¶ Learning rate VAE. [default: 1e-05]
-
-lr_mlp,--learning_rate_mlp<learning_rate_mlp>¶ Learning rate MLP. [default: 0.001]
-
-wd,--weight_decay<weight_decay>¶ Weight decay of adam optimizer. [default: 0.0001]
-
-dp,--dropout_p<dropout_p>¶ Dropout Percentage. [default: 0.2]
-
-e,--n_epochs<n_epochs>¶ Number of epochs to train over. [default: 50]
-
-s,--scheduler<scheduler>¶ Type of learning rate scheduler. [default: null]
-
-d,--decay<decay>¶ Learning rate scheduler decay for exp selection. [default: 0.5]
-
-t,--t_max<t_max>¶ Number of epochs before cosine learning rate restart. [default: 10]
-
-eta,--eta_min<eta_min>¶ Minimum cosine LR. [default: 1e-06]
-
-m,--t_mult<t_mult>¶ Multiply current restart period times this number given number of restarts. [default: 2.0]
-
-bs,--batch_size<batch_size>¶ Batch size. [default: 50]
-
-vp,--val_pkl<val_pkl>¶ Validation Set Methylation Array Location. [default: ./train_val_test_sets/val_methyl_array.pkl]
-
-w,--n_workers<n_workers>¶ Number of workers. [default: 9]
-
-v,--add_validation_set¶ Evaluate validation set.
-
-l,--loss_reduction<loss_reduction>¶ Type of reduction on loss function. [default: sum]
-
-hl,--hyperparameter_log<hyperparameter_log>¶ CSV file containing prior runs. [default: predictions/predict_hyperparameters_log.csv]
-
-j,--job_name<job_name>¶ Embedding job name. [default: predict_job]
-
-sft,--add_softmax¶ Add softmax for predicting probability distributions. Experimental.
regression_report¶
Generate regression report that gives concise results from regression tasks.
methylnet-predict regression_report [OPTIONS]
Options
-
-r,--results_pickle<results_pickle>¶ Results from training, validation, and testing. [default: predictions/results.p]
-
-o,--output_dir<output_dir>¶ Output directory. [default: results/]
methylnet-interpret¶
methylnet-interpret [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
bin_regression_shaps¶
Take aggregate individual scores from regression results, that normally are not categorized, and aggregate across new category created from continuous data.
methylnet-interpret bin_regression_shaps [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-t,--test_pkl<test_pkl>¶ Pickle containing testing set. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-c,--col<col>¶ Column to turn into bins. [default: age]
-
-n,--n_bins<n_bins>¶ Number of bins. [default: 10]
-
-ot,--output_test_pkl<output_test_pkl>¶ Binned shap pickle for further testing. [default: ./train_val_test_sets/test_methyl_array_shap_binned.pkl]
-
-os,--output_shap_pkl<output_shap_pkl>¶ Pickle containing top CpGs, binned phenotype. [default: ./interpretations/shapley_explanations/shapley_binned.p]
extract_methylation_array¶
Subset and write methylation array using top SHAP cpgs.
methylnet-interpret extract_methylation_array [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-co,--classes_only¶ Only take top CpGs from each class. [default: False]
-
-o,--output_dir<output_dir>¶ Output directory for methylation array. [default: ./interpretations/shapley_explanations/top_cpgs_extracted_methylarr/]
-
-t,--test_pkl<test_pkl>¶ Pickle containing testing set. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-c,--col<col>¶ Column to color for output csvs. [default: ]
-
-g,--global_vals¶ Only take top CpGs globally. [default: False]
-
-n,--n_extract<n_extract>¶ Number cpgs to extract. [default: 1000]
grab_lola_db_cache¶
Download core and extended LOLA databases for enrichment tests.
methylnet-interpret grab_lola_db_cache [OPTIONS]
Options
-
-o,--output_dir<output_dir>¶ Output directory for lola dbs. [default: ./lola_db/]
interpret_biology¶
Interrogate CpGs with high SHAPley scores for individuals, classes, or overall, for enrichments, genes, GO, KEGG, LOLA, overlap with popular cpg sets, GSEA, find nearby cpgs to top.
methylnet-interpret interpret_biology [OPTIONS]
Options
-
-a,--all_cpgs_pickle<all_cpgs_pickle>¶ List of all cpgs used in shapley analysis. [default: ./interpretations/shapley_explanations/all_cpgs.p]
-
-s,--shapley_data_list<shapley_data_list>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-o,--output_dir<output_dir>¶ Output directory for interpretations. [default: ./interpretations/biological_explanations/]
-
-w,--analysis<analysis>¶ Choose biological analysis. [default: GO]
-
-n,--n_workers<n_workers>¶ Number workers. [default: 8]
-
-l,--lola_db<lola_db>¶ LOLA region db. [default: ./lola_db/core/nm/t1/resources/regions/LOLACore/hg19/]
-
-i,--individuals<individuals>¶ Individuals to evaluate. [default: ]
-
-c,--classes<classes>¶ Classes to evaluate. [default: ]
-
-m,--max_gap<max_gap>¶ Genomic distance to search for nearby CpGs than found top cpgs shapleys. [default: 1000]
-
-ci,--class_intersect¶ Compute class shapleys by intersection of individuals. [default: False]
-
-lo,--length_output<length_output>¶ Number enriched terms to print. [default: 20]
-
-ss,--set_subtraction¶ Only consider CpGs relevant to particular class. [default: False]
-
-int,--intersection¶ CpGs common to all classes. [default: False]
-
-col,--collections<collections>¶ Lola collections. [default: ]
-
-gsea,--gsea_analyses<gsea_analyses>¶ Gene set enrichment analysis to choose from, if chosen will override other analysis options. http://software.broadinstitute.org/gsea/msigdb/collections.jsp [default: ]
-
-gsp,--gsea_pickle<gsea_pickle>¶ Gene set enrichment analysis to choose from. [default: ./data/gsea_collections.p]
-
-d,--depletion¶ Run depletion LOLA analysis instead of enrichment. [default: False]
-
-ov,--overlap_test¶ Run overlap test with IDOL library instead of all other tests. [default: False]
-
-cgs,--cpg_set<cpg_set>¶ Test for clock or IDOL enrichments. [default: IDOL]
-
-g,--top_global¶ Look at global top cpgs, overwrites classes and individuals. [default: False]
-
-ex,--extract_library¶ Extract a library of cpgs to subset future array for inspection or prediction tests. [default: False]
interpret_embedding_classes¶
Compare average distance between classes in embedding space.
methylnet-interpret interpret_embedding_classes [OPTIONS]
Options
-
-i,--embedding_methyl_array_pkl<embedding_methyl_array_pkl>¶ Use ./predictions/vae_mlp_methyl_arr.pkl or ./embeddings/vae_methyl_arr.pkl for vae interpretations. [default: ./embeddings/vae_methyl_arr.pkl]
-
-c,--pheno_col<pheno_col>¶ Column to separate on. [default: disease]
-
-m,--metric<metric>¶ Distance metric to compare classes. [default: cosine]
-
-t,--trim<trim>¶ Trim outlier distances. Number 0-0.5. [default: 0.0]
-
-o,--output_csv<output_csv>¶ Distances between classes. [default: ./results/class_embedding_differences.csv]
-
-op,--output_pval_csv<output_pval_csv>¶ If specify a CSV file, outputs pairwise manova tests between embeddings for clusters. [default: ]
list_classes¶
List classes/multioutput regression cell-types that have SHAPley data to be interrogated.
methylnet-interpret list_classes [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
list_individuals¶
List individuals that have ShapleyData. Not all made the cut from the test dataset.
methylnet-interpret list_individuals [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
order_results_by_col¶
Order results that produces some CSV by phenotype column, alphabetical ordering to show maybe that results group together. Plot using pymethyl-visualize.
methylnet-interpret order_results_by_col [OPTIONS]
Options
-
-i,--input_csv<input_csv>¶ Output directory for cpg jaccard_stats. [default: ./interpretations/shapley_explanations/top_cpgs_jaccard/all_jaccard.csv]
-
-t,--test_pkl<test_pkl>¶ Pickle containing testing set. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-c,--col<col>¶ Column to sort on. [default: disease]
-
-o,--output_csv<output_csv>¶ Output directory for cpg jaccard_stats. [default: ./interpretations/shapley_explanations/top_cpgs_jaccard/all_jaccard_sorted.csv]
-
-sym,--symmetric¶ Is symmetric? [default: False]
plot_all_lola_outputs¶
Iterate through all LOLA csv results and plot forest plots of all enrichment scores.
methylnet-interpret plot_all_lola_outputs [OPTIONS]
Options
-
-l,--head_lola_dir<head_lola_dir>¶ Location of lola output csvs. [default: interpretations/biological_explanations/]
-
-o,--plot_output_dir<plot_output_dir>¶ Output directory for interpretations. [default: ./interpretations/biological_explanations/lola_plots/]
-
-d,--description_col<description_col>¶ Description column for categorical variables [default: description]
-
-c,--cell_types<cell_types>¶ Cell types. [default: ]
-
-ov,--overwrite¶ Overwrite existing plots. [default: False]
plot_lola_output¶
Plot LOLA results via forest plot for one csv file.
methylnet-interpret plot_lola_output [OPTIONS]
Options
-
-l,--lola_csv<lola_csv>¶ Location of lola output csv. [default: ]
-
-o,--plot_output_dir<plot_output_dir>¶ Output directory for interpretations. [default: ./interpretations/biological_explanations/lola_plots/]
-
-d,--description_col<description_col>¶ Description column for categorical variables [default: description]
-
-c,--cell_types<cell_types>¶ Cell types. [default: ]
plot_top_cpgs¶
Plot the top cpgs locations in the genome using circos.
methylnet-interpret plot_top_cpgs [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-o,--output_dir<output_dir>¶ Output directory for output plots. [default: ./interpretations/output_plots/]
-
-i,--individuals<individuals>¶ Individuals to evaluate. [default: ]
-
-c,--classes<classes>¶ Classes to evaluate. [default: ]
produce_shapley_data¶
Explanations (coefficients for CpGs) for every individual prediction. Produce SHAPley scores for each CpG for each individualized prediction, and then aggregate them across coarser classes. Store CpGs with top SHAPley scores as well for quick access. Store in Shapley data object.
methylnet-interpret produce_shapley_data [OPTIONS]
Options
-
-i,--train_pkl<train_pkl>¶ Input database for beta and phenotype data. Use ./predictions/vae_mlp_methyl_arr.pkl or ./embeddings/vae_methyl_arr.pkl for vae interpretations. [default: ./train_val_test_sets/train_methyl_array.pkl]
-
-v,--val_pkl<val_pkl>¶ Val database for beta and phenotype data. Use ./predictions/vae_mlp_methyl_arr.pkl or ./embeddings/vae_methyl_arr.pkl for vae interpretations. [default: ./train_val_test_sets/val_methyl_array.pkl]
-
-t,--test_pkl<test_pkl>¶ Pickle containing testing set. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-m,--model_pickle<model_pickle>¶ Pytorch model containing forward_predict method. [default: ./predictions/output_model.p]
-
-w,--n_workers<n_workers>¶ Number of workers. [default: 9]
-
-bs,--batch_size<batch_size>¶ Batch size. [default: 512]
-
-c,--cuda¶ Use GPUs.
-
-ns,--n_samples<n_samples>¶ Number of samples for SHAP output. [default: 200]
-
-nf,--n_top_features<n_top_features>¶ Top features to select for shap outputs. If feature selection, instead use this number to choose number features that make up top global score. [default: 20]
-
-o,--output_dir<output_dir>¶ Output directory for interpretations. [default: ./interpretations/shapley_explanations/]
-
-mth,--method<method>¶ Explainer type. [default: kernel]
-
-ssbs,--shap_sample_batch_size<shap_sample_batch_size>¶ Break up shapley computations into batches. Set to 0 for only 1 batch. [default: 0]
-
-r,--n_random_representative<n_random_representative>¶ Number of representative samples to choose for background selection. Is this number for regression models, or this number per class for classification problems. [default: 0]
-
-col,--interest_col<interest_col>¶ Column of interest for sample selection for explainer training. [default: disease]
-
-rt,--n_random_representative_test<n_random_representative_test>¶ Number of representative samples to choose for test set. Is this number for regression models, or this number per class for classification problems. [default: 0]
-
-e,--categorical_encoder<categorical_encoder>¶ One hot encoder if categorical model. If path exists, then return top positive controbutions per samples of that class. Encoded values must be of sample class as interest_col. [default: ./predictions/one_hot_encoder.p]
-
-cc,--cross_class¶ Find importance of features for prediction across classes.
-
-fs,--feature_selection¶ Perform feature selection using top global SHAP scores. [default: False]
-
-top,--top_outputs<top_outputs>¶ Get shapley values for fewer outputs if feature selection. [default: 0]
-
-vae,--vae_interpret¶ Use model to get shapley values for VAE latent dimensions, only works if proper datasets are set. [default: False]
-
-cl,--pred_class<pred_class>¶ Prediction class top cpgs. [default: ]
-
-r,--results_csv<results_csv>¶ Remove all misclassifications. [default: ./predictions/results.csv]
-
-ind,--individual<individual>¶ One individual top cpgs. [default: ]
-
-rc,--residual_cutoff<residual_cutoff>¶ Activate for regression interpretations. Standard deviation in residuals before removing. [default: 0.0]
-
-cn,--cell_names<cell_names>¶ Multioutput names for multi-output regression. [default: ]
-
-dsd,--dump_shap_data¶ Dump raw SHAP data to numpy file, warning, may be large file. [default: False]
reduce_top_cpgs¶
Reduce set of top cpgs.
methylnet-interpret reduce_top_cpgs [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-nf,--n_top_features<n_top_features>¶ Top features to select for shap outputs. [default: 500]
-
-o,--output_pkl<output_pkl>¶ Pickle containing top CpGs, reduced number. [default: ./interpretations/shapley_explanations/shapley_reduced_data.p]
regenerate_top_cpgs¶
Increase size of Top CpGs using the original SHAP scores.
methylnet-interpret regenerate_top_cpgs [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-nf,--n_top_features<n_top_features>¶ Top features to select for shap outputs. [default: 500]
-
-o,--output_pkl<output_pkl>¶ Pickle containing top CpGs, reduced number. [default: ./interpretations/shapley_explanations/shapley_reduced_data.p]
-
-a,--abs_val¶ Top CpGs found using absolute value.
-
-n,--neg_val¶ Return top CpGs that are making negative contributions.
return_shap_values¶
Return matrix of shapley values per class, with option to, classes/individuals are columns, CpGs are rows, option to plot multiple histograms/density plots.
methylnet-interpret return_shap_values [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-o,--output_dir<output_dir>¶ Output directory for output plots. [default: ./interpretations/shap_outputs/]
-
-i,--individuals<individuals>¶ Individuals to evaluate. [default: ]
-
-c,--classes<classes>¶ Classes to evaluate. [default: ]
-
-hist,--output_histogram¶ Whether to output a histogram for each class/individual of their SHAP scores.
-
-abs,--absolute¶ Use sums of absolute values in making computations.
-
-log,--log_transform¶ Log transform final results for plotting.
shapley_jaccard¶
Plot Shapley Jaccard Similarity Matrix to demonstrate sharing of Top CpGs between classes/groupings of individuals.
methylnet-interpret shapley_jaccard [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-c,--class_names<class_names>¶ Class names. [default: ]
-
-o,--output_dir<output_dir>¶ Output directory for cpg jaccard_stats. [default: ./interpretations/shapley_explanations/top_cpgs_jaccard/]
-
-ov,--overall¶ Output overall similarity. [default: False]
-
-i,--include_individuals¶ Output individuals. [default: False]
-
-co,--cooccurrence¶ Output cooccurrence instead jaccard. [default: False]
-
-opt,--optimize_n_cpgs¶ Search for number of top CpGs to use. [default: False]
split_hyper_hypo_methylation¶
Split SHAPleyData object by methylation type (needs to change; but low methylation is 0.5 beta value and below) and output to file.
methylnet-interpret split_hyper_hypo_methylation [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-o,--output_dir<output_dir>¶ Output directory for hypo/hyper shap data. [default: ./interpretations/shapley_explanations/shapley_data_by_methylation/]
-
-t,--test_pkl<test_pkl>¶ Pickle containing testing set. [default: ./train_val_test_sets/test_methyl_array.pkl]
-
-thr,--threshold<threshold>¶ Pickle containing testing set. [default: original]
view_methylation_top_cpgs¶
Write the Top CpGs for each class/individuals to file along with methylation information.
methylnet-interpret view_methylation_top_cpgs [OPTIONS]
Options
-
-s,--shapley_data<shapley_data>¶ Pickle containing top CpGs. [default: ./interpretations/shapley_explanations/shapley_data.p]
-
-i,--individuals<individuals>¶ Individuals. [default: ]
-
-o,--output_dir<output_dir>¶ Output directory for cpg methylation shap. [default: ./interpretations/shapley_explanations/top_cpgs_methylation/]
-
-t,--test_pkl<test_pkl>¶ Pickle containing testing set. [default: ./train_val_test_sets/test_methyl_array.pkl]
methylnet-visualize¶
methylnet-visualize [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
plot_roc_curve¶
Plot ROC Curves from classification tasks; requires classification_report be run first.
methylnet-visualize plot_roc_curve [OPTIONS]
Options
-
-r,--roc_curve_csv<roc_curve_csv>¶ Weighted ROC Curve. [default: results/Weighted_ROC.csv]
-
-o,--outputfilename<outputfilename>¶ Output image. [default: results/roc_curve.png]
plot_training_curve¶
Plot training curves as output from either the VAE training or VAE MLP.
methylnet-visualize plot_training_curve [OPTIONS]
Options
-
-t,--training_curve_file<training_curve_file>¶ Training and validation loss and learning curves. [default: predictions/training_val_curve.p]
-
-o,--outputfilename<outputfilename>¶ Output image. [default: results/training_curve.png]
-
-vae,--vae_train¶ Plot VAE Training curves.
-
-thr,--threshold<threshold>¶ Loss values get rid of greater than this.
methylnet-torque¶
methylnet-torque [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
run_torque_job¶
Run torque job.
methylnet-torque run_torque_job [OPTIONS]
Options
-
-c,--command<command>¶ Command to execute through torque. [default: ]
-
-gpu,--use_gpu¶ Specify whether to use GPUs. [default: False]
-
-a,--additions<additions>¶ Additional commands to add, one liner for now. [default: ]
-
-q,--queue<queue>¶ Queue for torque submission, gpuq also a good one if using GPUs. [default: default]
-
-t,--time<time>¶ Walltime in hours for job. [default: 1]
-
-n,--ngpu<ngpu>¶ Number of gpus to request. [default: 0]